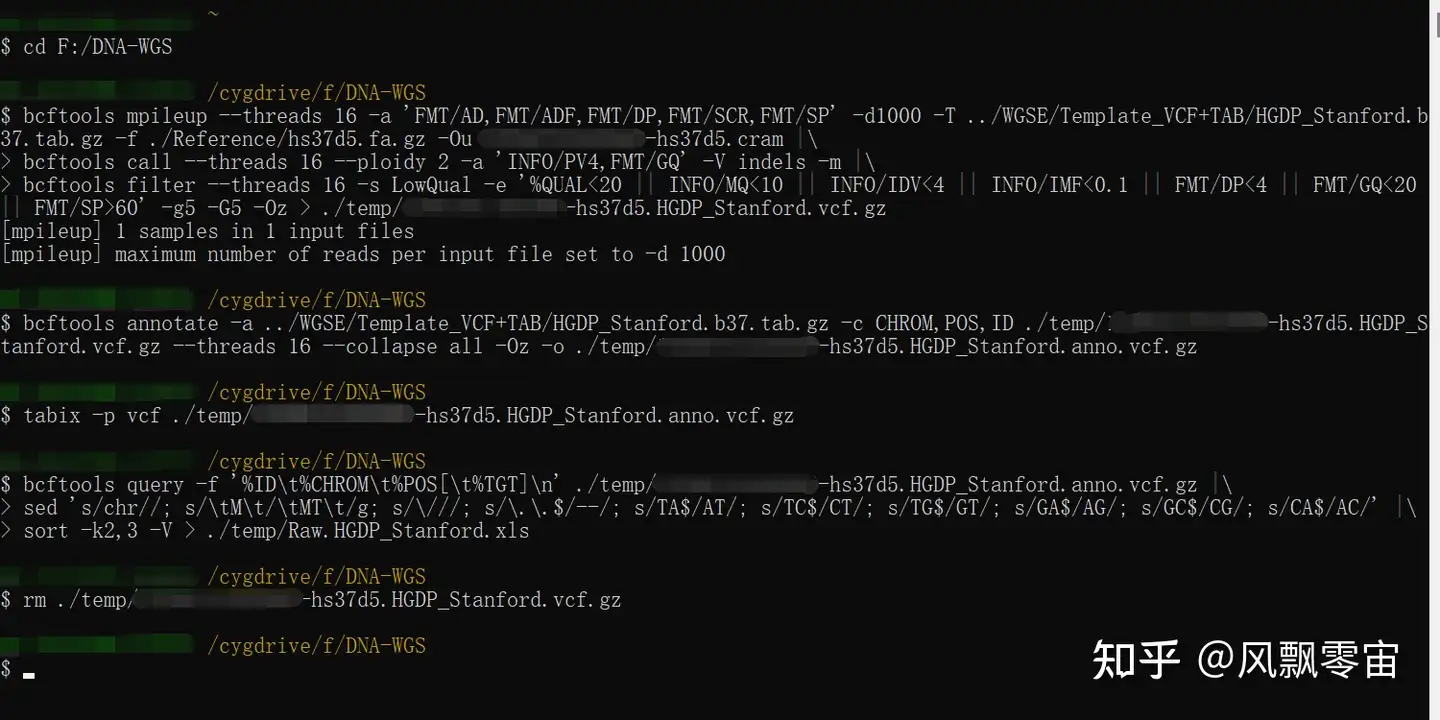
Protocol for unbiased, consolidated variant calling from whole exome sequencing data - ScienceDirect

bcftools view | bcftools tutorial on how to count the number of snps and indels in a vcf file - YouTube
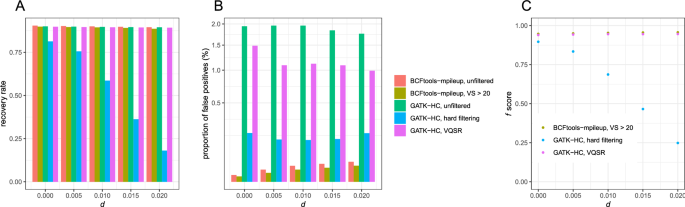
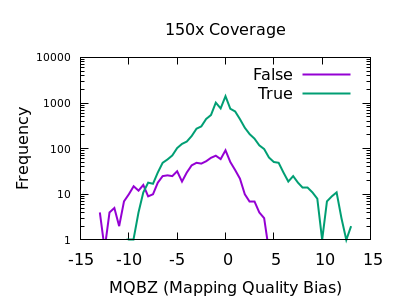
The evaluation of Bcftools mpileup and GATK HaplotypeCaller for variant calling in non-human species | Scientific Reports
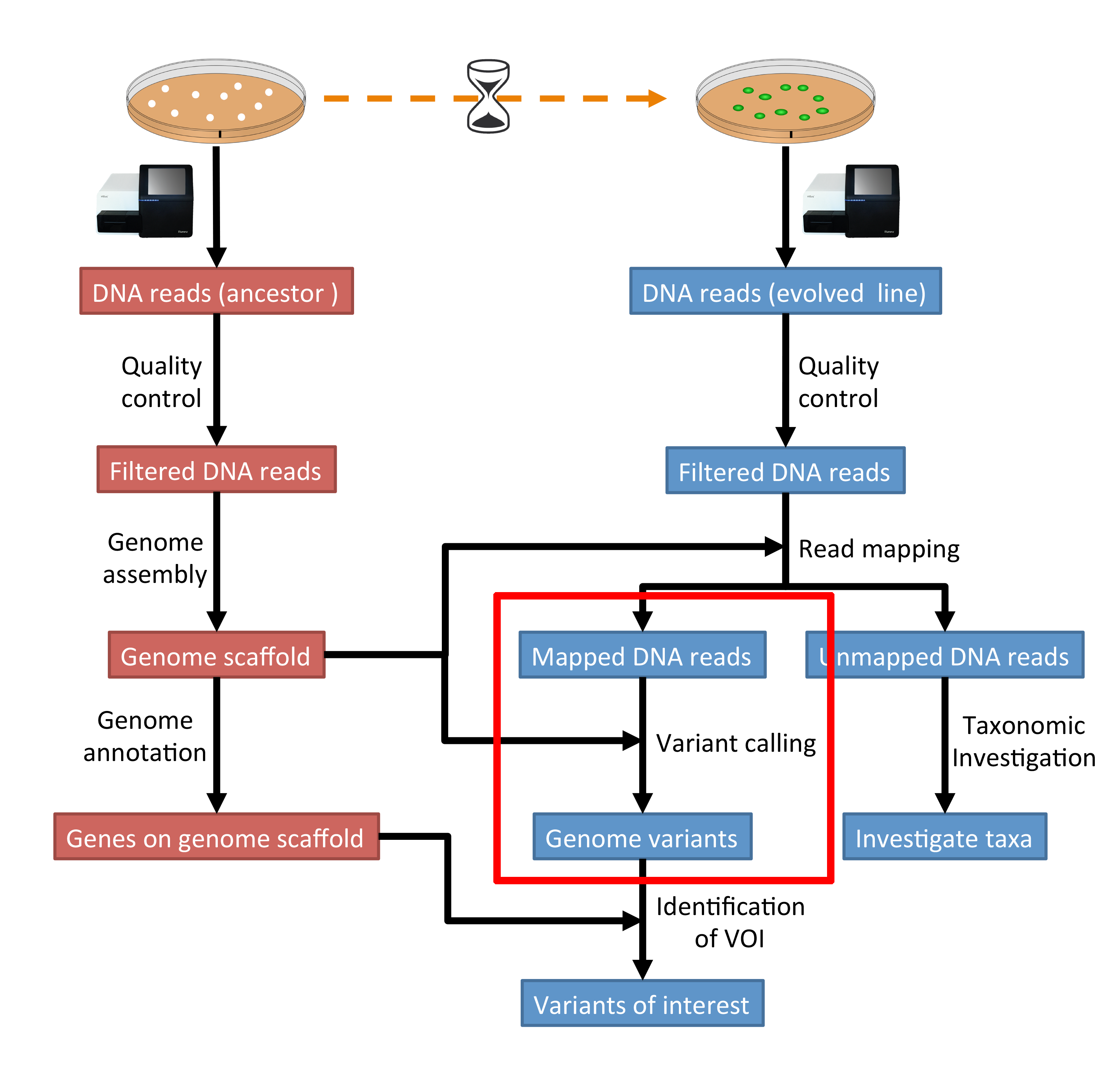
No unexpected CRISPR-Cas9 off-target activity revealed by trio sequencing of gene-edited mice | PLOS Genetics

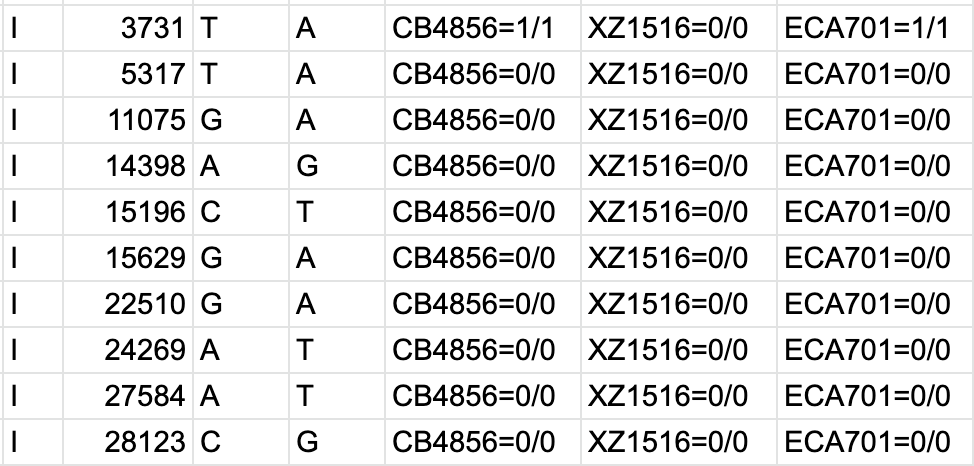
bcftools filter -e 'GT="het"' recognize GT 2/2 as heterozygous · Issue #1268 · samtools/bcftools · GitHub